Pia Vogel
Professor
Biological Sciences
| Office Location |
DLSB 233 |
| Phone |
214-768-1790 |
Education
Ph.D. University of KaiserlauternLab: DLSB 221
Tel: 214-768-1785
Email: pvogel@smu.edu
Research Interests
Structure, Molecular Dynamics and Protein-Protein Interactions of Nucleotide Binding Proteins
F1Fo -ATP-Synthase
Multidrug Resistance Proteins
Ryanodine Receptor
ATP Synthase: Protein-Protein Interactions
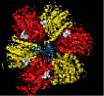

I have studied F1-ATPase for more than 20 years using a number of different approaches. Since a structure for the mitochondrial F1-ATPase was solved in 1994 by John Walker's laboratory and the rotational motion of the internal γ-subunit of the enzyme can be observed in real time in the experiments from Masasuke Yoshida's laboratory, the questions that are still intriguing and unresolved center around the
My group is focusing on the interactions and the structure of the so-called external stalk of the ATPsynthase that is thought to stabilize the enzyme complex and may be involved in elastic energy coupling between the proton-driven rotation of membrane embedded subunits of the enzyme and the rotation of the internal γ-subunit that causes the binding and release of substrate (ADP) and product (ATP).
Using the powerful Electron Spin Resonance Spectroscopy (ESR), we employ site-specific spin labeling of either the whole enzyme or the potential "stator-subunit" b to elucidate structural and dynamic information. Heavy use is made of state-of-the-art molecular modeling techniques. The data collected in the lab are used to validate structural models created on our two Linux computer clusters.
Multi-Drug Resistance: Regulation, Structure and Function
The family of ABC-type ATPases performs a wide variety of transport processes across cellular membranes. Transport substrates range from import-substrates like nutrients, e.g. sugars, ions and 
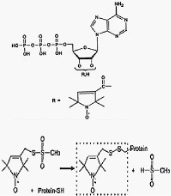
Although high-resolution structures exist for some of the bacterial transporters, much remains to be learned about their molecular mechanisms and the protein dynamics that are key to the enzyme mechanisms. Such information will be extremely valuable even if an X-ray structural model became available in the near future for the human protein. In our group we perform a variety of highly innovative experiments using ESR spectroscopy combined with spin-labeled nucleotides and site-specific spin labeling to further our understanding of the molecular mechanisms that underlie the function of this medically greatly relevant enzyme. We currently study the human drug resistance proteins MDR1 and the MRPs 2 and 3.
Ryanodine Receptor: Structure and Function
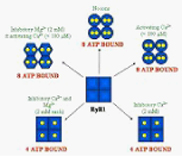
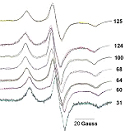
We have recently discovered a potentially novel pathway for the regulation of calcium channels like the ryanodine sensitive Ca-channel, ryanodine receptor 1 (RyR1), using Electron Spin Resonance spectroscopy (ESR) and spin-labeled ATP. Employing this technique we for the first time unequivocally showed that the homo-tetrameric channel contains a total of eight ATP-binding sites and that the accessibility and affinity of these binding sites is directly regulated by divalent cations like Mg2+ and Ca2+.
Scientific Accomplishments

Selected Publications
“Subunit b dimer of the Escherichia coli ATP synthase can form left-handed coiled coils”, John G. Wise and Pia D. Vogel, (2007) submitted
“Structure of the Cytosolic Part of the Subunit b-Dimer of Escherichia coli FoF1-ATP Synthase”, Tassilo Hornung, Oleg A. Volkov, Tarek M. A. Zaida, Sabine Delannoy, John G. Wise and Pia D. Vogel, (2007) submitted
“Insights into the Regulation of the Ryanodine Receptor: Differential Effects of Mg2+ and Ca2+ on Nucleotide Binding”, Dias, J.M., Szegedi, C, Jóna, I., and Vogel, P.D. (2006) Biochemistry 45, 9408 – 9415
